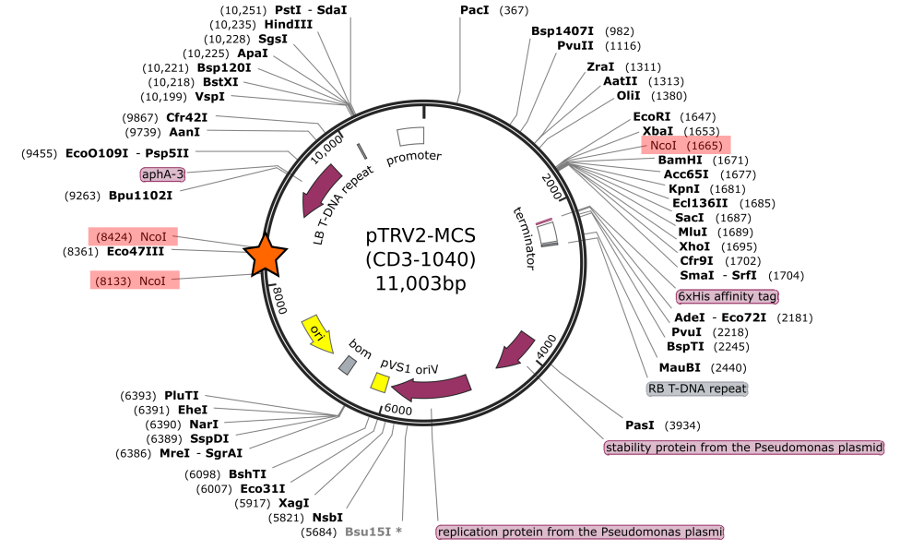
I recently began cloning constructs to perform VIGS experiments on flowering plants. I ordered the pTRV2-MCS vector from the Arabidospsis biological resource center ABRC which is a vector that supports cloning a target fragment into a modified Tobacco Rattle Virus. I based the design of my cloning primers on the vector map and sequence available from the ordering page. According to that map, there is a single Nco1 restriction site (CCATGG) and a single Bamh1 site (GGATCC) in the multiple cloning site (MCS). Having had success with these enzymes in the past I chose to use them for my cloning project. So, I designed a forward primer with the Nco1 site and a reverse primer with the Bamh1 and proceded to begin the cloning process. However, when I digested the pTRV2 vector with Nco1 and Bamh1 an ~300bp band appeared on my agarose gel below the linearized vector band. There are supposed to be only a few bases between these two restriction sites, so obviously something was wrong!
I contacted the ABRC to ask about this issue. The very nice people from ABRC got back to me quickly and sent along a revised sequence of the vector (see map graphic below) along with some additional information. I was surprised to learn that in fact there is an insertion of an E. coli genomic region into the pTRV2-MCS vector. The only indication of this is a note at the bottom of the order page that reads “The vector contains a 1kb insert, which doesn’t affect the function. The donor is currently creating a replacement version of the vector.” However, it is unclear from this note where this insertion is or how it changes the vector. It is clear from looking at the revised vector map that this insertion introduces two additional Nco1 sites, with an approximately 300bp region between them, which explains the results of my RE digest. Obviously, I felt uncomfortable continuing attempts at cloning with this combination of enzymes, so I switched to Xba1 for the forward primer (I also confirmed via digest of the vector that this combination does not yield any unexpected bands!).
Suffice it to say, take care when cloning into the pTRV2-MCS vector! And, if you’re ordering a vector such as I did, I recommend taking extra precautions to double-check that everything behaves as you expect. I hope that this post saves others from a cloning headache.
Revised vector map of CD3-1040 pTRV2-MCS vector (sequence is here.)
Insertion is marked by orange star. Extra Nco1 sites are highlighted in red.