Working at the interface of biochemistry and evolutionary biology
Wordcloud generated from the abstracts of all my published papers

Over the course of my undergraduate and graduate careers my interests have shifted progressivley more toward evolutionary biology. Evolution is the driving force that underlies all of the biological complexity and diversity on Earth. That a random process—governed solely by the physical rules of the universe—can give rise to amazingly complex, self-organizing, self-replicating systems is both beautiful and amazing. What are the mechanisms of this process? How are simple physical and chemical principles translated into the amazing complexity that we see in living systems? How does the architecture of individual living things constrain evolutionary outcomes? How do interactions within populations shape evolutionary processes? These are the types of questions that drive my passion for scientific research!
Hybridization, phenotypic evolution, and pollination in Penstemon
My work with Dr. Carrie Wessinger at the University of South Carolina aimed to characterize the history and adaptive impacts of hybridization and introgression in the Habroanthus section of the genus Penstemon. We combined field studies in natural hybrid zones, whole-genome re-sequencing, phylogenomics, transcriptomics, and phenotypic measurements to understand how introgression between species with different pollinators has influenced the evolution of plant phenotypes. The goal was to determine whether adaptive introgression was involved in the repeated evolution of the hummingbird pollinated phenotype in Habroanthus and how different pollination systems have shaped the population-level dynamics and macroevolutionary history in the clade. I have continued working on independent projects to characterize the pollinator community for several Penstemon species native to Colorado.
The molecular mechanisms of flower color evolution

My work with Dr. Stacey Smith is focused on understanding the mechanisms that underly evolutionary transitions in flower color. Flower color plays an essential role in the biology and ecology of angiosperms. Evolution of novel flower colors is often associated with adaptation to new environments or pollinators. The pathways underlying this polygenic phenotype are relatively well characterized, making it an excellent system to probe molecular mechanisms and address broader evolutionary questions. How are evolutionary trajectories in polygenic phenotypes constrained? What are the relative contributions of regulatory and functional changes? How do phenomena such as epistasis and pleiotropy affect the predictability of evolution? By reconstructing historical evolutionary transitions between flower colors in the family Solanaceae and determining the genetic and biochemical bases of these changes, we have gained important insights into how changes in pathway genes shape the evolution of floral pigmentation phenotypes. To tackle this complex problem I have used a diverse array of approaches including transcriptomics, phylogenetics, biochemical characterization, and computational modeling.
The evolutionary biochemistry of the S100 protein family
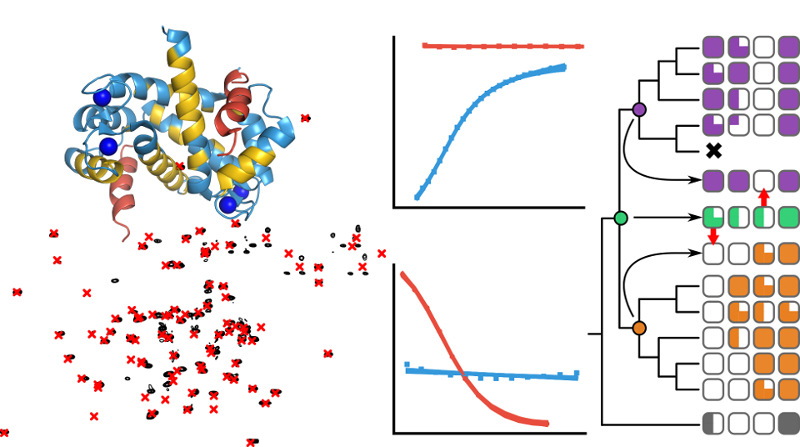
My PhD research in the Harms lab at University of Oregon was centered around understanding how the physical and chemical properties of proteins shape evolutionary outcomes. I focused on two main case studies as a way of probing several important principles in protein evolution; 1) The evolutionary divergence of metal binding sites in the S100 protein family as a model to understand the evolutionary lability of biochemical mechanisms, and 2) The evolution of binding specificity in the S100s as a model to study changes in specificity following gene duplication. My work took an interdisciplinary approach using a variety of tools from the fields of biochemistry, molecular biophysics, evolutionary biology, and molecular biology. Joining together experimental techniques such as phage display, next-generation sequencing, and isothermal titration calorimetry with computational techniques like ancestral sequence reconstruction and machine learning opened up the door to gaining deep insights into evolutionary stories that would otherwise be inaccessible.
