My work in the Harms lab at University of Oregon focused on understanding the evolution of protein biochemical properties and activities. My later work, comprising the last chapters of my dissertation, was centered on the evolution of peptide binding specificity in the S100 protein family. Using molecular phylogenetics and ancestral sequence reconstruction (ASR) we were able to trace the evolutionary shifts in specificity within a particular clade of the S100 family. Finally, the last chapter of my dissertation is available in the form of two bioRxiv pre-prints. Both articles are also currently in review at society journals.
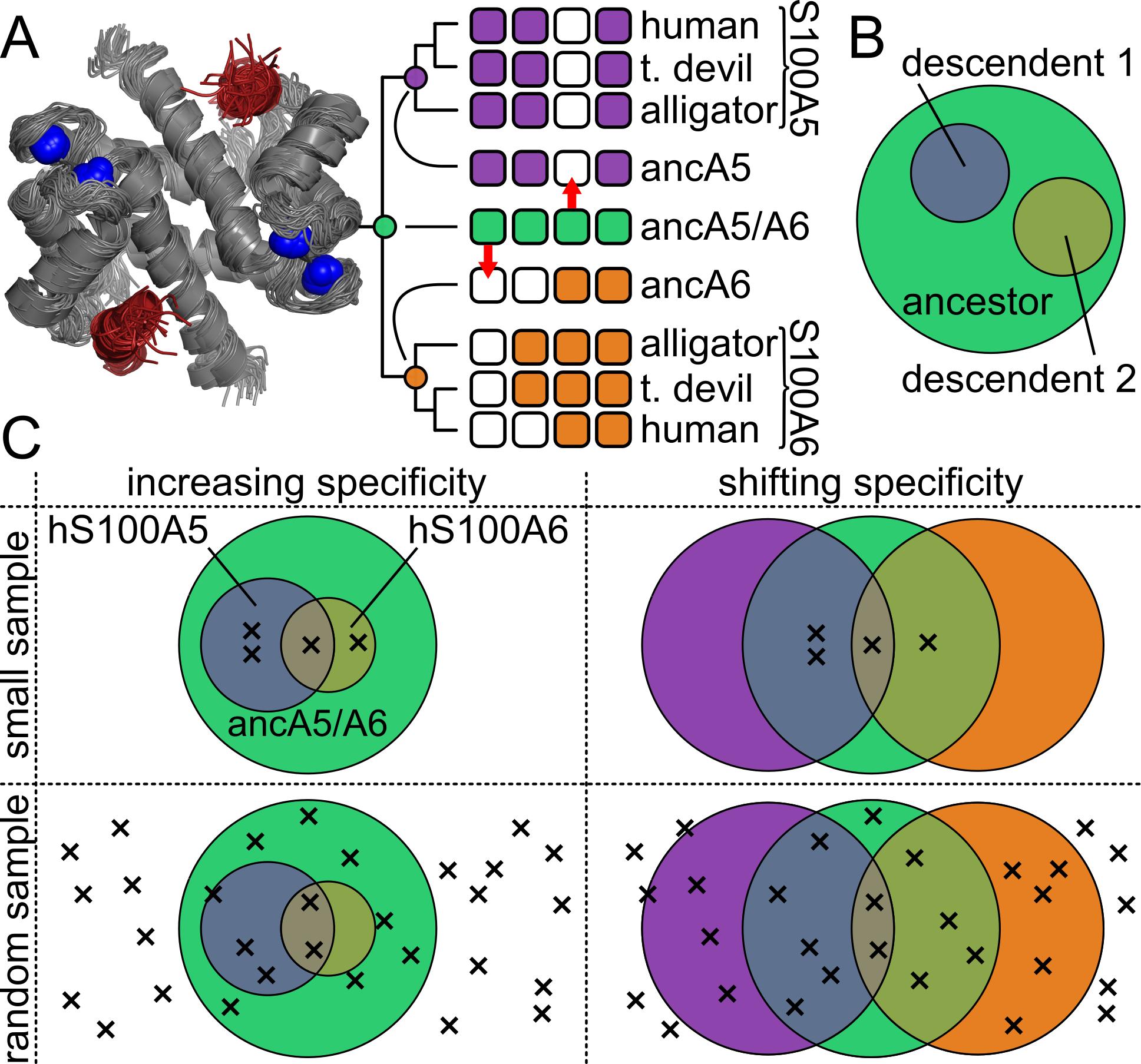
In the first study we used a combination of high-throughput phage display binding screens and ASR to quantitatively examine shifts in the intrinsic binding specificity of S100 proteins for their peptide targets. We found that this historical pattern does not conform the to hypothesis that specificity of proteins generally increases over the course of evolution, instead supporting a more nuanced set of shifts that differ between protein lineages.
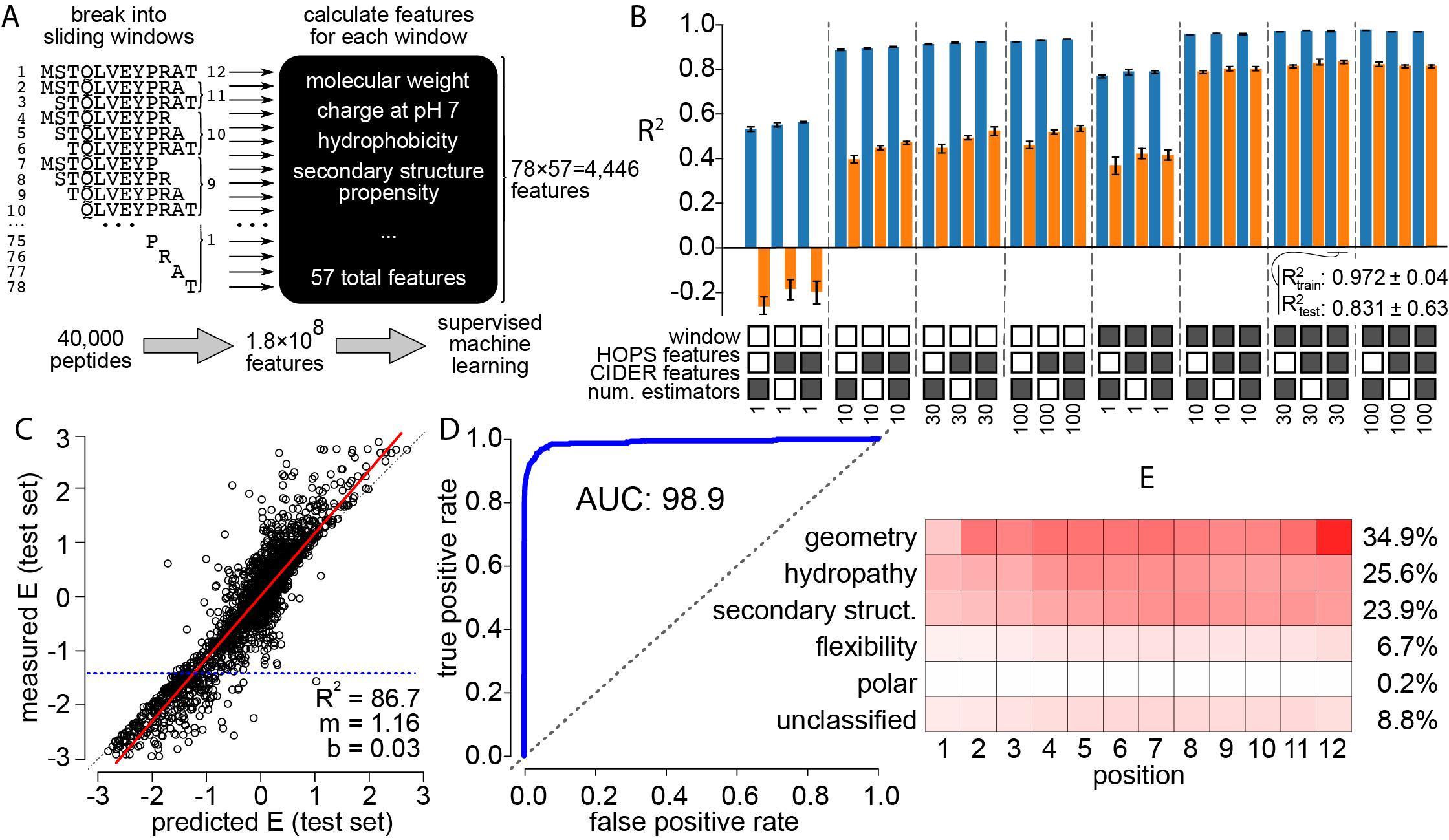
The second pre-print focuses on the protein S100A5. We used a machine-learning approach, in combination with docking simulations and x-ray crystallography to identify the key biochemical properties that predict peptide binding. S100s are notorious for having “low specificity” that is essentially impossible to summarize using traditional motif-based concepts. We find that a set of biochemical features of peptides, such as hydrophobicity, shape complementarity, and secondary structure propensity can be used to accurately predict phage-display enrichment. These results are further explained by solving a crystal structure of S100A5 and modeling peptide binding to the protein surface.