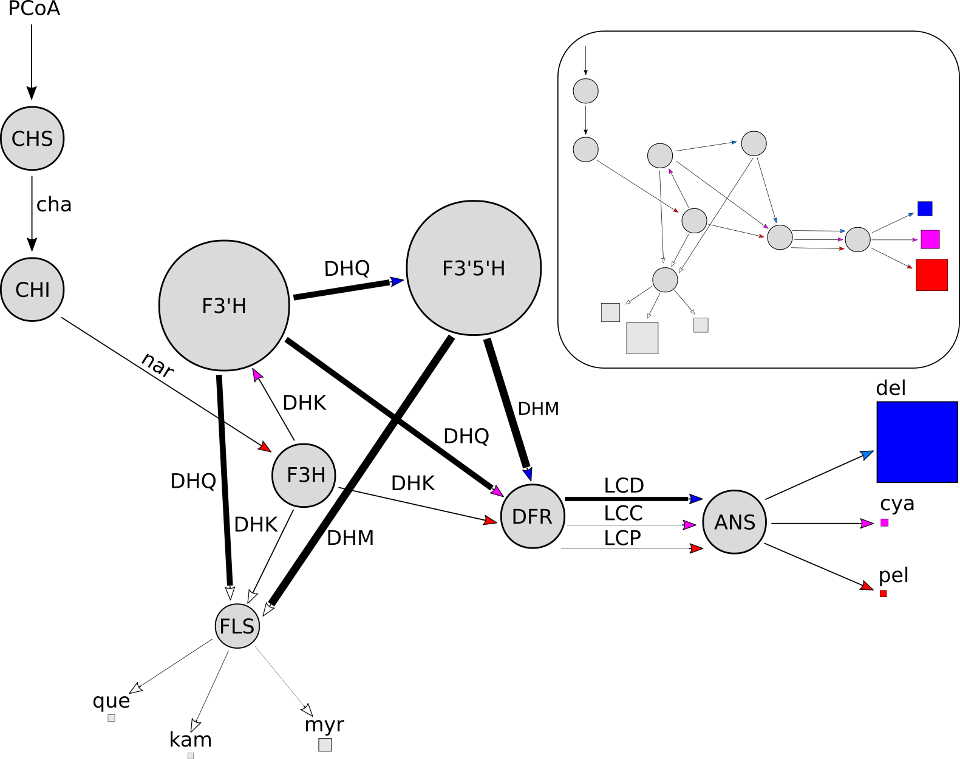
My work in the Smith lab at CU Boulder is focused on understanding the molecular mechanisms of flower color evolution. One of the primary biochemical pathways responsible for the production of colorful pigments is the anthocyanin pathway. Mutations to enzymes in this pathway have been repeatedly observed to be responsible for shifts in floral pigmentation. However, there are not enough empirical observations, at this point in time, to draw quantitative conclusions regarding the distribution of mutations across pathway genes, the distribution of types of mutations, and the underlying constraints that give rise to patterns.
To help inform our expectations and understand the intrinsic constraints imposed by anthocyanin pathway structure, I developed a mathematical model describing pathway dynamics. I then developed an evolutionary simulation framework (availabe on github) that allows us to computationally evolve the pathway model between defined pigmentation phenotypes. By conducting about ten thousand parallel simulations we were able to study the statistical properties of evolutionary trajectories. We discovered that 1) the metabolic control structure of the pathway shifts to facilitate changes in pigment production, 2) certain pathway enzyme genes, located at key branch points, are the primary targets of selection for shifting pigmentation, and 3) there are strong and predictable trade-offs between branches in the pathway that constrain evolutionary trajectories.
We previously made this work available as a pre-print on bioRxiv and I am happy to announce that the work has also just been published online in Integrative and Comparative Biology.
The simulated data generated from this study, along with the scripts used to conduct the simulations and the Jupyter notebook used to conduct our analyses, can be downloaded from Zenodo.